BIOE Associate Professor Explores How Huntington’s Protein Detects Curved Membranes
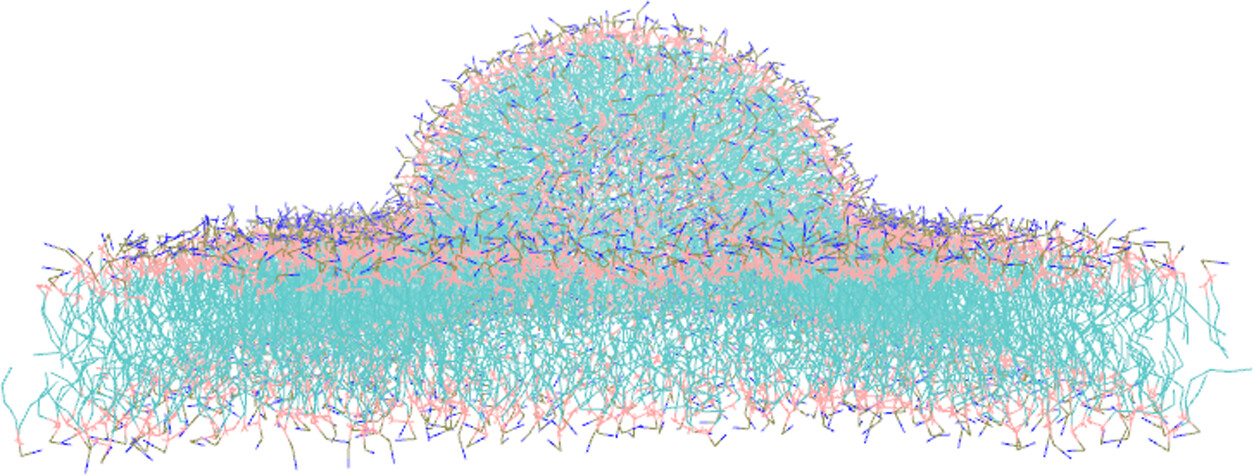
Huntington’s disease begins with changes in a protein found in every cell of the body, called huntingtin. In people with the disease, a section of this protein is longer than usual, causing it to misfold and form harmful clusters inside nerve cells. A short region at the front of the protein, known as Nt17, plays an essential role in this process. Because this small segment influences early disease events, understanding how Nt17 behaves is key to identifying where Huntington’s disease starts at the molecular level. A new study led by Fischell Department of Bioengineering (BIOE) Associate Professor Silvina Matysiak reveals how a portion of the huntingtin protein recognizes and binds to curved lipid membranes. This research, published in ACS Chemical Neuroscience, offers new insight into how membrane interactions may shape the early stages of Huntington’s disease.
In the study, Matysiak and her collaborators combined computer simulations with experimental measurements to show that Nt17 binds more readily to curved membrane regions than to flat ones. Two phenylalanine amino acids at the ends of Nt17 drive this effect by fitting into spots on the membrane where lipids are loosely packed. This behavior may explain why Nt17 clusters at specific cellular sites and how these early interactions promote harmful protein aggregation by increasing local protein concentration. In the lab’s simulations, wild-type Nt17 molecules attached preferentially to curved membrane regions. When the phenylalanine residues were replaced with smaller hydrophobic ones, curvature sensitivity dropped sharply. Experimental tests confirmed that Nt17 bound more strongly to small, highly curved vesicles than to larger, flatter ones. “Targeting how Nt17 interacts with membrane curvature gives us a new direction for Huntington’s therapies focusing on how this peptide engages with lipid membrane surfaces inside cells,” Matysiak explains. These findings shed light on how fragments of the huntingtin protein gather at particular cellular locations, where they can trigger damaging aggregation. Understanding this binding step could inform efforts to prevent or slow disease progression.
The paper suggests that targeting Nt17’s membrane interactions, rather than focusing solely on huntingtin’s polyglutamine expansion, could open new therapeutic possibilities. Strategies that mask the phenylalanine residues or modulate membrane properties may reduce the protein’s tendency to aggregate. As principal investigator of the Biomolecular Modeling Lab, Matysiak and her team specialize in connecting molecular behavior with broader biological effects. The lab develops multiscale simulations to study how environmental heterogeneity, like membrane interfaces with distinct chemical and physical properties, macromolecular crowding, and polysaccharides, influences protein shapes, how proteins assemble, and how they localize on surfaces. This research supports their broader goal of linking molecular mechanisms to disease outcomes. The work was conducted in collaboration with Computational Biology PhD candidate Neha Nanajkar, UMD alumnus Abhilash Sahoo, and Chemistry & Biochemistry Professor Shelli Frey at Gettysburg College, and was supported by the National Science Foundation and UMD Supercomputing resources. With its clear demonstration of how a small protein segment detects and shapes membrane environments, this study brings scientists closer to understanding the molecular origins of Huntington’s disease and other disorders linked to protein misfolding.
Related Articles: November 18, 2025 Prev Next |
|