BCE Students Use Nanopore Sequencing to Connect Biology and Data Science
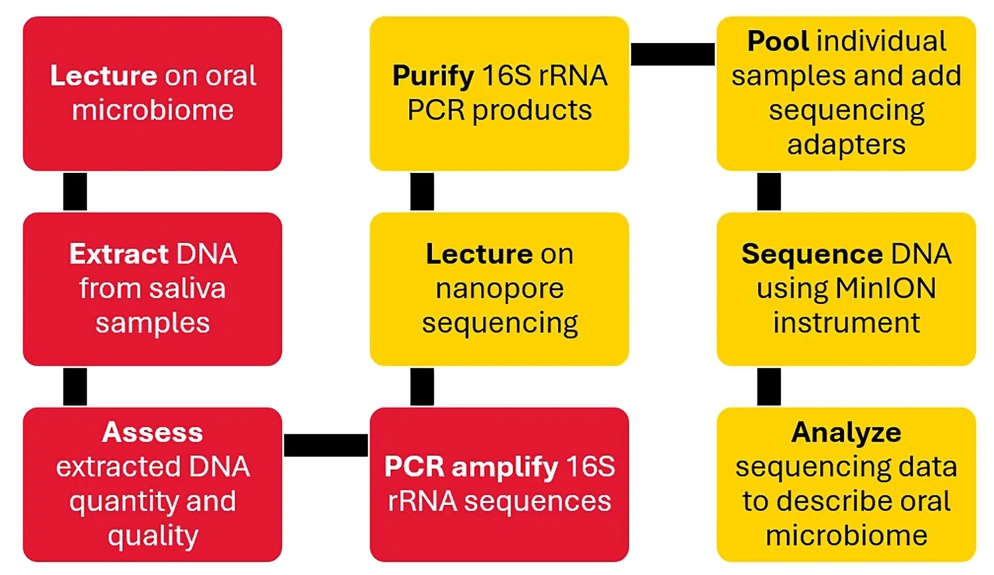
Students at the Fischell Department of Bioengineering’s Biocomputational Engineering (BCE) program are gaining hands-on training in DNA sequencing through a new lab module that blends wet-lab biology with advanced data analysis. The module’s findings were recently published in Biomedical Engineering Education. The two-week module was designed by BCE Lecturer Jarred Callura, as part of the program’s Molecular Techniques Laboratory. In this activity, students extract DNA from their own saliva samples, prepare sequencing libraries, and analyze the microbial communities that comprise their oral microbiomes. “I am constantly looking for ways to improve our BCE courses to enrich our students' learning experiences, and our entire program takes student feedback seriously,” Callura said. “It was actually discussions with the first BCE graduates that sparked the creation of the nanopore sequencing module as a way to improve the lab course.” The approach paid off as students who were typically more focused on coding and computation successfully performed complex molecular biology techniques. Sequencing runs produced high-quality data, with one semester generating more than 100 gigabases and another producing over a million reads in less than five hours. These results confirmed that BCE students are capable of mastering demanding lab skills while producing datasets that are valuable for computational analysis.
“The combination of wet-lab work with the generation and analysis of huge datasets exemplifies the unique advantages of Biocomputational Engineering as compared to computer science and traditional bioengineering,” Callura said. “Our students gain a diverse set of skills that will help them be successful in any biotech or data science career they choose.” Students stated the module gave them a stronger sense of how biology and computation work together. Many appreciated being able to take part in every stage of the process, from collecting samples to analyzing the data, which helped them see the bigger picture of how lab work ties into research. Others highlighted how using their own saliva samples made the activity more meaningful. Working with personal data, rather than an anonymous specimen, made the lab feel more engaging and memorable. “My favorite part of the lab was learning about nanopore sequencing technology and deepening my understanding of bioinformatics analysis. Also, it was quite interesting to see the different microbes present in my mouth,” one student said.
Related Articles: September 26, 2025 Prev Next |