AI Tool Reveals Gaps in Ancestry Reporting Across Biomedical Research
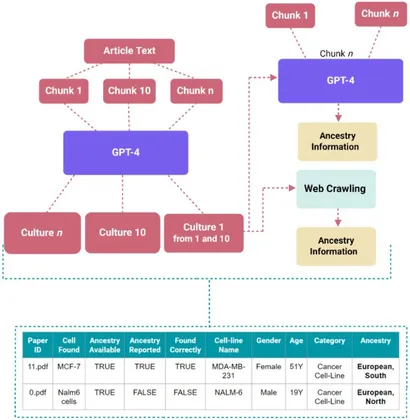
Researchers from the University of Maryland’s Fischell Department of Bioengineering (BIOE) have developed an artificial intelligence tool that showcases how often biomedical studies fail to report ancestry information in their research samples. Beyond identifying these gaps, the system helps chart a path toward more accurate and comprehensive biomedical science. The system, called TRACE (Tool for Researching Ancestry and Cell Extraction), offers a data-driven way to evaluate representation in preclinical science. This research, led by BIOE researcher Alison M. Veintimilla in Dr. Erika Moore’s lab, along with colleagues at the University of Florida, was published this month in Frontiers in Digital Health. “We’re so excited to share this work and the AI tool that will enable us to extract more information about how it is considered in which diseases and aspects of biomedical research,” says Erika Moore. Ancestry is one of the most significant, yet often overlooked, variables in biomedical research. Cell lines and tissue samples form the foundation of countless experiments that are used to model disease, test drugs, and understand genetic risk. However, most of these samples originate from a narrow subset of populations, typically of European descent. This lack of variation can influence how results translate to different communities and may contribute to disproportionate outcomes in medicine. By identifying where these imbalances occur, researchers can begin to design studies that better reflect real-world human variation. To address this issue, Veintimilla and her colleagues built TRACE, a large-scale screening tool powered by natural language processing and data mining. The program scans scientific articles to identify mentions of human cell lines or primary tissue samples, then compares those references against public databases to determine whether ancestry is recorded and what populations are represented. By automating this process, TRACE enables scientists to evaluate thousands of publications in a fraction of the time it would take to do so manually.
When applied across a curated collection of biomedical studies, TRACE revealed evident trends. In many papers, ancestry information was completely missing, and when it was available, most cell lines could be traced back to populations of European ancestry. These findings reinforce earlier concerns that biomedical data may not accurately reflect global human variation. “I see this research as a step 0.5 in a much larger process,” said Veintimilla. “TRACE helps establish a baseline for how ancestry is currently reported so we can build stronger systems for data accuracy and reliability in the future.” The researchers also discovered surprising findings related to how ancestry is described in scientific writing. Authors use inconsistent language—sometimes listing geographic regions, other times ethnic groups or population codes—making it difficult for both humans and AI systems to categorize results. TRACE was built to recognize and reconcile these inconsistencies, providing a standardized framework for future analyses. The team emphasized that AI tools alone cannot resolve issues in research. Because language models can misinterpret or “hallucinate” information, human validation remains an essential step in ensuring the accuracy of outputs. Still, by combining automation with expert review, TRACE makes it a practical tool for large-scale evaluation of scientific trends. “Computational models are powerful, but they depend on the quality of the data they are given,” Veintimilla said. “By pairing automation with human oversight, we can make meaningful progress toward more representative science.” Beyond its technical advances, TRACE showcases how inclusivity is measured in the laboratory. The open-access code and database, available through GitHub, allow other researchers to test and expand the tool for their own work.
Related Articles: October 21, 2025 Prev Next |
|